MAJIQ Simplifier
MAJIQ and VOILA have been important tools in the mRNA/RNA Seq world since they were released in 2016. One of the most characteristic and relevant features that MAJIQ provides is the ability to detect and quantify denovo and complex splicing variations. A side effect of this is that it can become very complex very quickly, especially when you start running large datasets detecting all the new variations that where not defined before.
Another aspect that needs to be considered is how trustworthy an annotation DB is. In model organisms such as human or mouse, the transcripts in the annotation DB are often miss-annotated.
What is MAJIQ Simplifier?
The MAJIQ --simplify option is a new argument during the builder step that removes non-relevant the splicing variations. Simplifying away non-relevant splicing variations makes it easier for the user to understand the biology. This simplification is done based on read ratio between junctions (default) and also raw read counts per junction (optional).
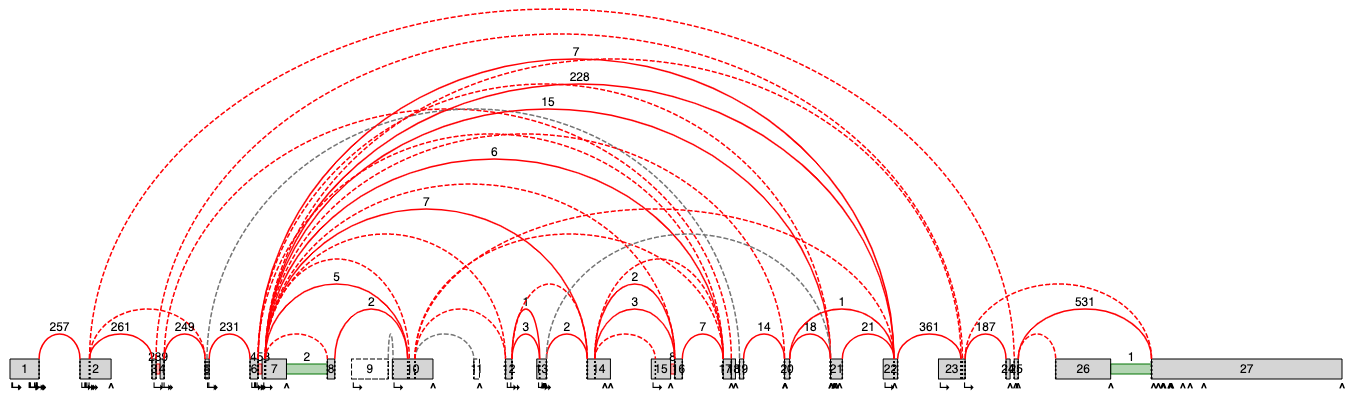
In this example image the high number of junctions is polluting the splice graph making it highly
difficult to analyze.
Most of these junctions are annotated, but seldom utilzied (read ratio less than 1% in over half the
samples in all of the the builder groups).
For MAJIQ, these junctions are real and trustworthy, but they are not relevant for this study, so
--simplify discards them from any lsv.
From now on we will call such discarded junctions, irrelevant junctions.
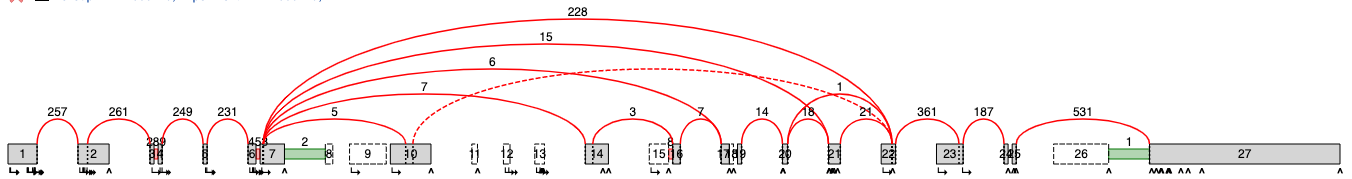
After simplifying, the splicegraph looks cleaner and clearer for a posterior analysis.
How to use MAJIQ Simplifier?
MAJIQ simplifier is a part of the builder pipeline, that is triggered using the argument --simplify . This
argument enables the simplification process that can be tweaked using a set of specific parameters.
--simplify [psi_threshold]: Simplify enables the simplification step, the ratio used to simplify is specified by psi_threshold, 0.01 by default.--simplify-denovo readnum: Simplify all denovo junctions which total number of raw reads is lower than readnum. Default value is 0--simplify-annotated readnum: Simplify all annotated junctions which total number of raw reads is lower than readnum. Default value is 0--simplify-ir readnum: Simplify all intron retentions which total number of reads is lower than readnum. Default value is 0
Only --simplify argument is required to activate the simplification, in which case the rest of the arguments would be set to their default values.
What happens to the LSVs?
After the simplification steps, all the LSVs are defined and quantified without considering the irrelevant junctions. Their future quantification would not be possible without re-running the builder step without simplifier or with other thresholds.
Simplifier effects in VOILA
VOILA visualization is mostly the same by default. The effects of the simplification are visible using a new button than can be activated or deactivated to show the splicegraph structure with or without the irrelevant junctions. You can see the effect in the animation below.
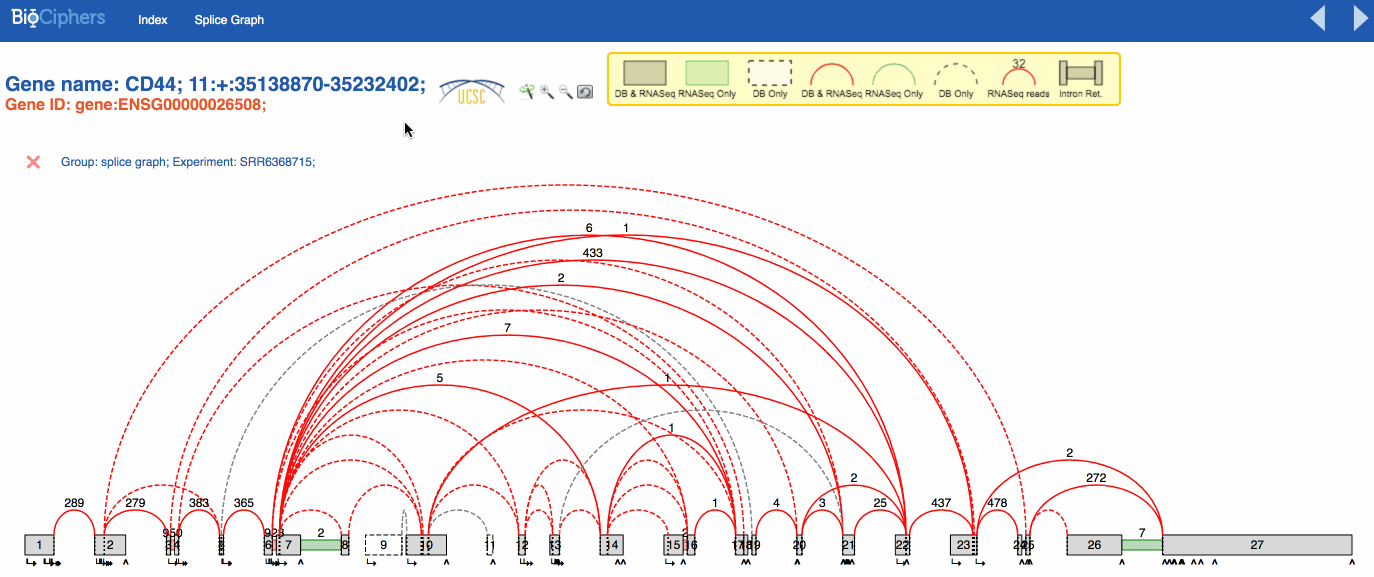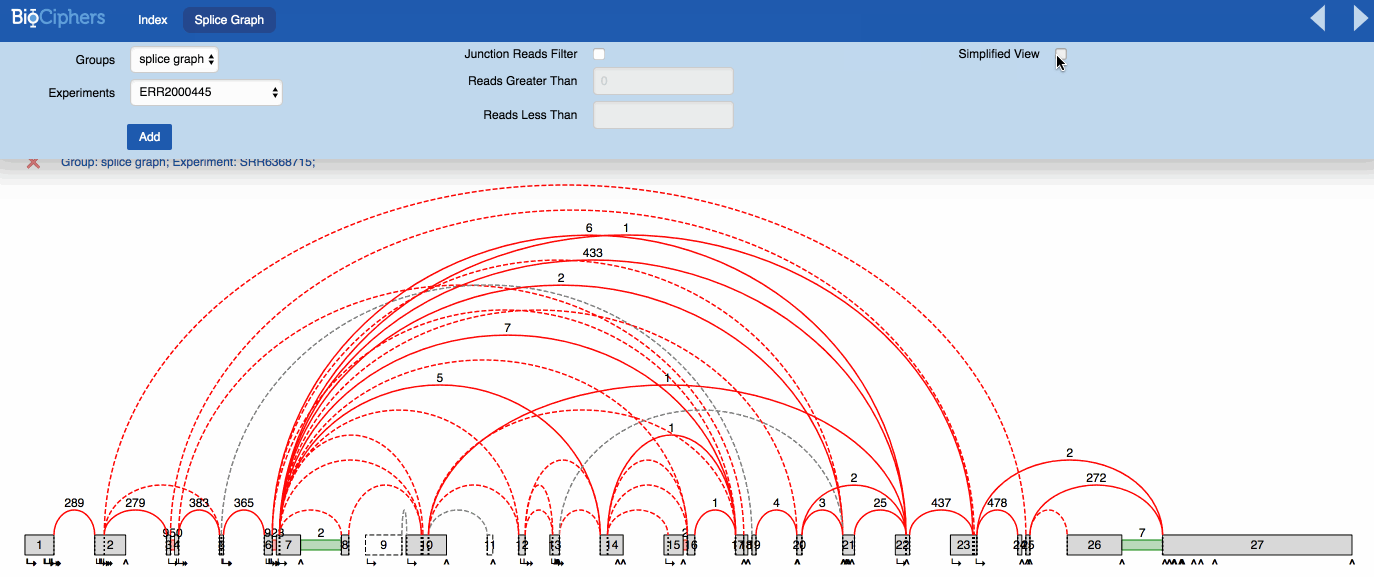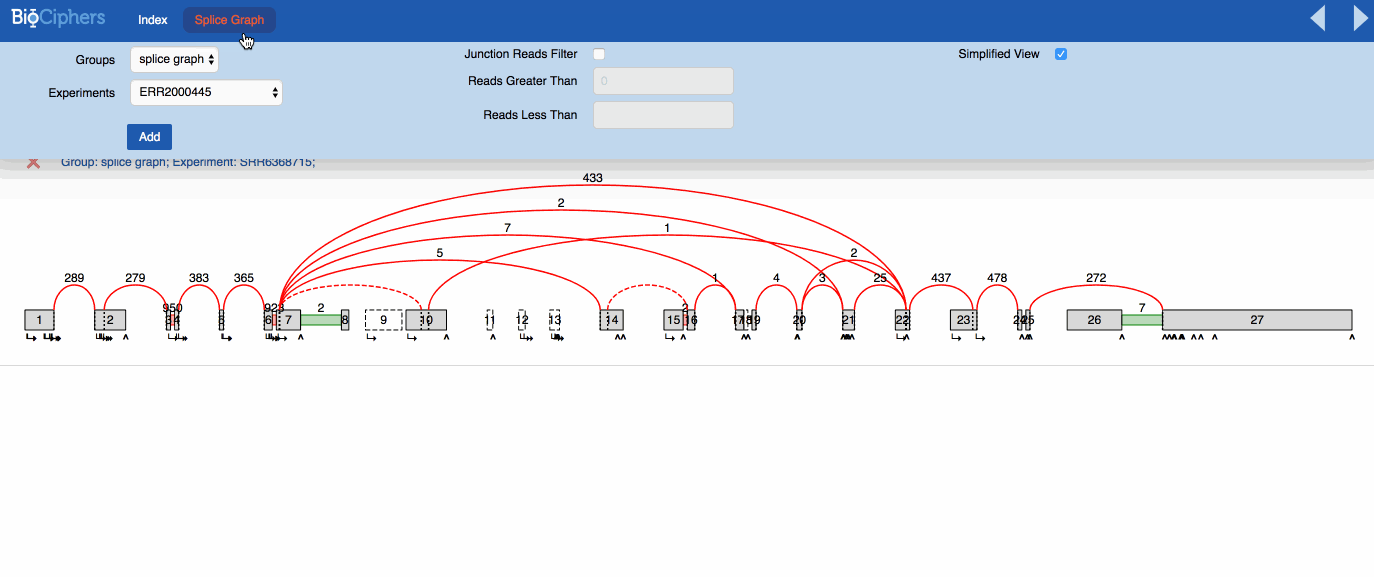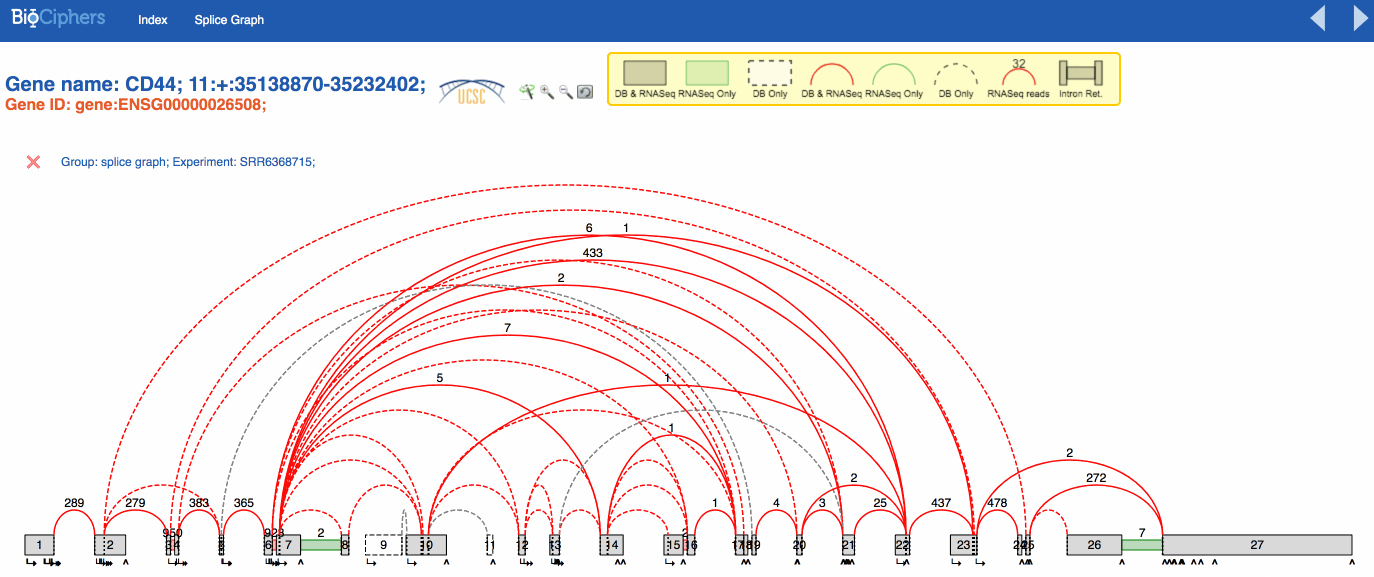
NOTE: The LSVs only show the quantification over simplified LSVs, the new button only affects the splicegraph.